APP tune and run APPS
The APP Tune dialog used for tuning and running APPs. APP Tune allows for easy image analysis without knowledge about the underlying classification method, pre- and post processing steps, etc. Once the APP has been set up, APP Tune will suffice for the image analysis. This is especially applicable when several users are using the software but only one authority can edit the details of the protocol.
The default classifier in Visiopharm is the deep learning classifier, which can be tuned in the APP Author, under the "tune" tab. Other classifiers can be tuned via "Advanced Sampling".
In this article, a brief description is provided on how to tune APPs in the APP Tune dialog using the Threshold classifier. If APP Tune is opened with an APP that uses a classifier other than Threshold, classification parameters cannot be changed or tuned. However, APP Tune can still be used to preview and run APPs with the output calculations.
APP Tune is contextual, meaning that the dialog changes depending on which type of classifier that the current APP uses.
Tune APPs with threshold classification
An example of the Threshold classifier is seen below with and without added image classes for training and output.
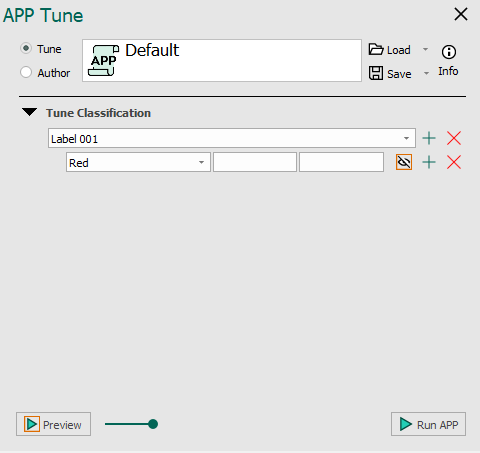
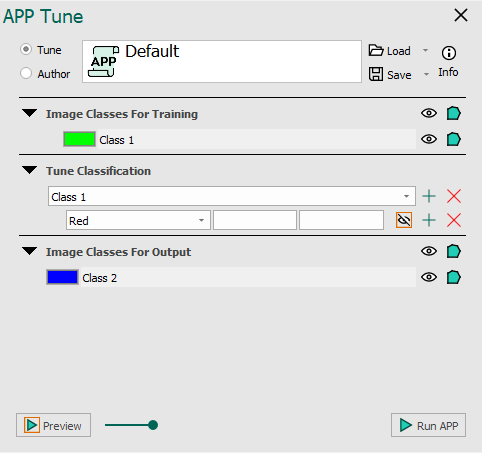
The APP tune author for the Threshold classifier. No classes added for training seen on the left and classes added for training seen on the right.
Once an APP has been configured to use the Threshold classifier in APP Author, it may need to be tuned for a specific image or application. This is done under Tune Classification, where the threshold value used in the classification can be specified. Choose the lower and upper limits for the current label and feature. The limits should be specified as pixel values in the respective feature image.
Tune APPs with cell classification
All Cell Classification APPs use a texture-based classifier different from the rest of the classifiers in Visiopharm. This also effects the appearance of APP Tune dialog. All configuration options for the classifier are located in APP Tune, whereas the other classifiers of Visiopharm are configured in APP Author.
For APPs using Cell Classification as the classification method, it is possible to complete advanced image analysis problems without prior knowledge of image analysis techniques. The use of APP Tune is not complicated compared to the complexity of the problem and the quality of the results. If an APP has been set up (either locally or included as a standard APP from Visiopharm), only a few decisions have to be made by the user.
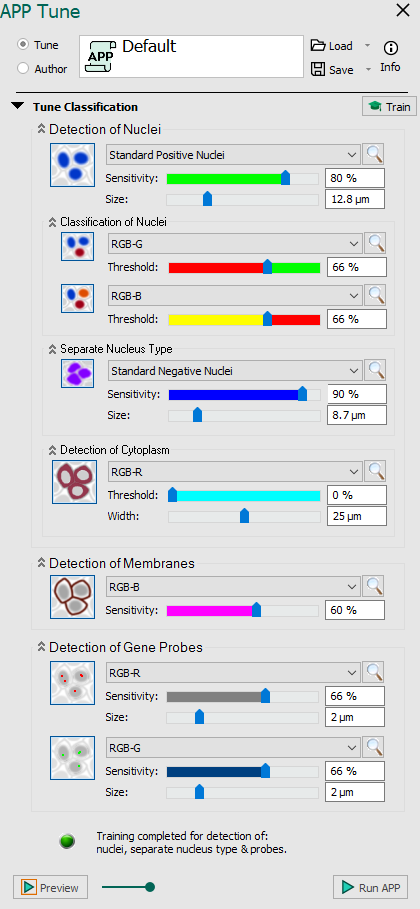
| Parameter | Description |
|---|---|
 | Enable detection of nuclei Detect nuclei based on feature image using Label 001. Move the slide bars to see the effect of different levels sensitivity and size. The nuclei detection functionality is based on two dictionaries build from small image patches of intensity and label data. The small image patches are randomly sampled from a ROI where all nuclei have been assigned a label by the user. From the dictionaries, a probability map is constructed. The probability map holds probabilities of the given pixel being a nucleus pixel. By thresholding the probability map, the algorithm is able to classify background and nuclei. The image is segmented by going through all the pixels in the input image and extracting an image patch. The image patch is then classified by considering the proximity to similar patches in the intensity and label dictionaries. |
 | Enable classification of detected nuclei This function provides the option for dividing the detected nuclei into two groups using Label 003. The function classifies nuclei as positive vs. negative based on a feature image (e.g. stain separated image). Adjust the threshold to see the effect. By considering the mean stain intensity of nuclei in a given input band, the algorithm is able to separate individual nucleus based on whether their stain intensity is above or below the mean value. The separation can be adjusted with a threshold parameter. |
 | Enable secondary classification of detected nuclei This function provides the option for further subdivision of the stained nuclei using Label 004. Adjust the level of positivity manually and see the effect. |
 | Enable detection of separate nucleus type Train the classifier to detect another type of nuclear if two types of nuclei with distinct shapes are present in the image. Uses Label 002 for training. Move the slide bars to see the effect of different levels sensitivity and size. This options is always enabled if the feature Standard Positive Nuclei is selected under Detection of Nuclei. As an extension to the nuclei detection algorithm, the separate nucleus type can detect two types of nuclei instead of one. This is useful for detecting two nuclei markers or when the size and texture of one type of nuclei vary to such an extent that it is hard to capture the variations using only one class of nuclei. Note that this differs from the nuclei classification in that the separate nucleus type detects two different types of nuclei from the beginning, which can then be further separated into several classes using the nuclei classification functionality. |
 | Enable detection of cytoplasm Detect cytoplasm corresponding to all detected nuclei based on a feature image. Move the slider bars to adjust the threshold for detection and width. Cytoplasm is outlined by Label 006, when the value is above threshold and by Label 007 for values below. Does not need any training. Due to uneven distribution and staining of the cytoplasm, it is often very difficult to find the accurate delineation of the cytoplasm. Here we define an area surrounding each nucleus as cytoplasm from the nucleus border. Despite the limitation of not detecting the real cytoplasm, it is possible to extract meaningful features from the cytoplasm region. |
 | Enable detection of membranes Train the texture classifier for detection of membranes. Manually choose the sensitivity of detection by moving the slider bar or entering a percentage. Membranes are outlined by Label 005. Does not need any training, but requires minimum 20x magnification. The cell membrane is often expressed as linear structures surrounding the nuclei. Local linear structures are localized by polynomial filtering and approximated by mean filtering a skeletonization. |
 | Enable detection of first probe type Detect Gene Probes based on feature image using Label 008. Move the slide bars to see the effect of different levels sensitivity and size. Gene probes are fragments of DNA or RNA that are used to detect complementary gene targets. In histological images, the probes are often seen as colorized dots or blobs at the location. The probe detection functionality uses the same algorithm as for the nuclei detection. |
 | Enable detection of second probe type This function provides the option for detecting a second Gen Probe type using Label 009. Adjust the threshold to see the effect. |
| Train detection Click this option to train the classifier with a user-defined example. This will only work if the feature images have label(s) and an ROI(s). | |
  | Indicator Indicates whether or not the training is completed and the processing can begin. |