Working with Layer Data
Layer Data (ROIs, Annotations, Labels and Heatmaps) of the active image can be saved to a file and imported to other images, or processed in other programs.
Alternatively, the Regions Of Interest (ROIs) can be extracted separately to an .xml
Export Layer Data
-
Click on
 or on
or on  .
. -
Navigate to and select the image with the layer data to export.
-
Click File | Export | Layer Data to start the export process.
-
Browse to the location where the file should be saved and give it an appropriate name. Select the export type and click Save to export the layer data in the selected format.
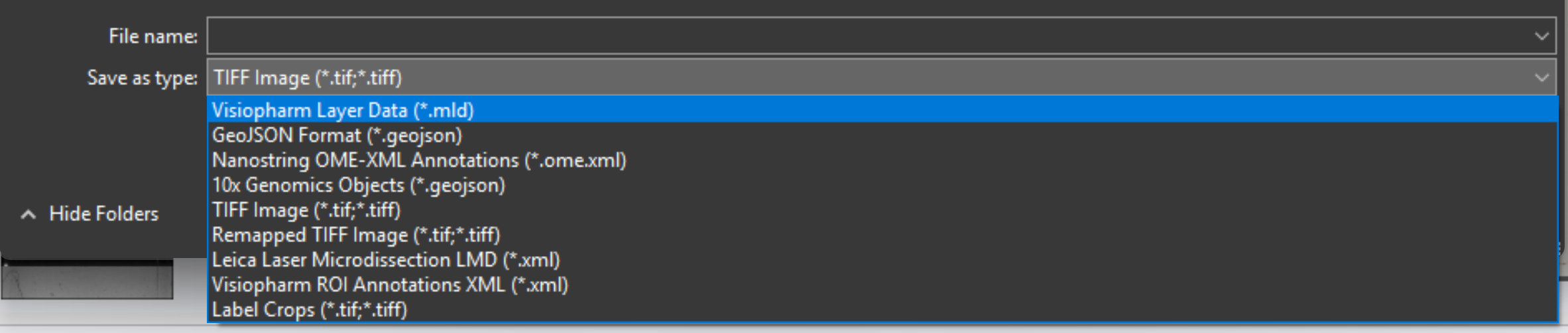
Supported Export Formats
The following file formats are available for export in Visiopharm:
| File Type | Description | Extension |
|---|---|---|
| Visiopharm Layer Data | Contains data layers created in Visiopharm for analysis or visualization. | .mld |
| GeoJSON format | Export labels in geojson format compatible with Xenium Eplorer software. | .geojson |
| Nanostring OME-XML Annotations | Exported annotations in XML format compatible with Nanostring OME analysis workflows. | .ome.xml |
| TIFF image | High-resolution image format commonly used in microscopy and imaging software. | .tif .tiff |
| Remapped TIFF image | Used to save overlays in the reference frame of a linked Tissuealign image. It will create the same output as TIFF in a single image. | .tif .tiff |
| Leica Laser Microdissection LMD | Exported ROI annotations in XML format compatible with Leica LMD software. | .xml |
| Visiopharm ROI Annotations XML | Region of interest annotations exported as XML files for integration with other systems. | .xml |
| Label Crops | Used for import of whole slide 32-bit TIFFs with segmentation or annotation data, and convert the objects into Visiopharm Labels. Used when the TIFF image comes with 32-bit integer values (intensity levels) with uniquely IDed objects. All objects in the imported TIFF file will be converted to overlay Labels of type 1, and objects are split with NN4 connectivity. 0 intensity will give no Label. | .tif .tiff |
GeoJSON for 10X Genomics Xenium Export
Labels created in the Visiopharm software can be exported in the .geojson format for further analysis in Xenium Explorer. Please see https://www.10xgenomics.com/support/software/xenium-ranger/latest/analysis/running-pipelines/XR-import-segmentation for more details.
Only label objects are exported and non-solid labels (that is, those with any holes) will be excluded upon import. For each label object, an Object ID is written both to the GeoJSON file and the "Object Info" output variable which allows correlation of the Visiopharm and Xenium objects after import.
Remapped TIFF image Export
The Remapped TIFF image format is used to save overlays in the reference frame of a linked Tissuealign image. It will create the same output as TIF in a single image. Tissuealign workflows are detailed in the section Tissuealign. One usage is exemplified in the following analysis where a slide is scanned over two points in time, and each scan is slightly misaligned due to being moved in and out of the scanner. The analysis has been made on a reference slide denoted Section 1, resulting in an ROI around the tissue. If this is exported as layer data in a standard TIFF image format. When importing the layer data from Section 1 onto a later scan of the same slide, denoted Section 2, the layer data is misaligned due to the misalignment between the Section 1 and Section 2 scans.
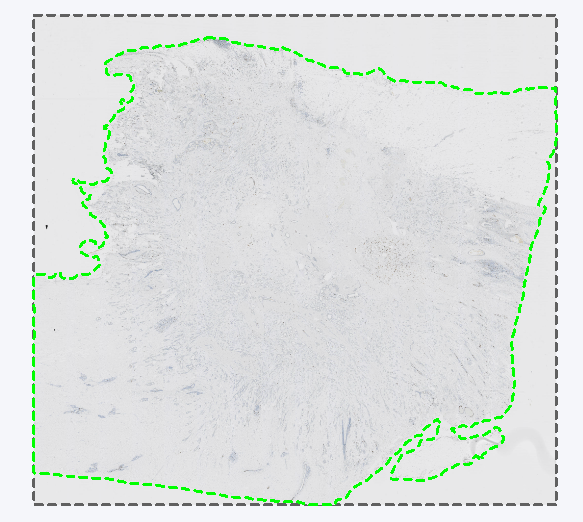
Section 1 with a ROI around the tissue exported as layer data in TIFF format.
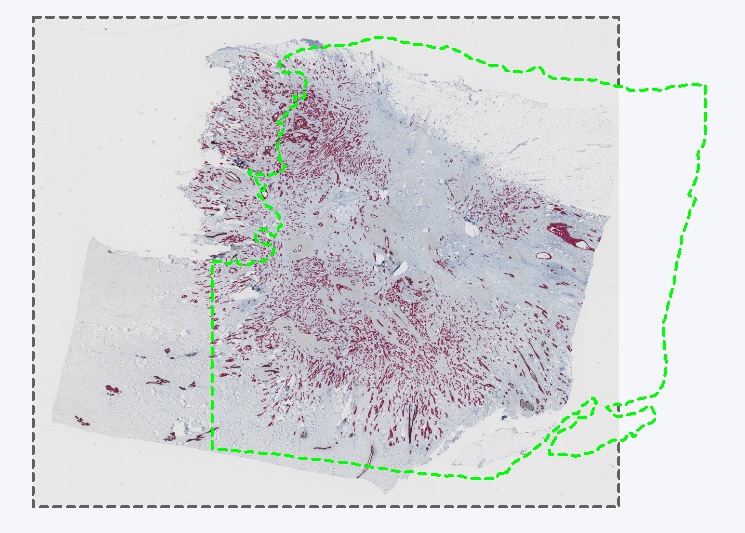
Section 2 with the misaligned layer data from the Section 1 slide.
In such cases one would go through a Tissuealign workflow, or potentially merely auto align the Section 1 and Section 2 scans, to align the layer data and continue with the analysis on the Section 2 scan.
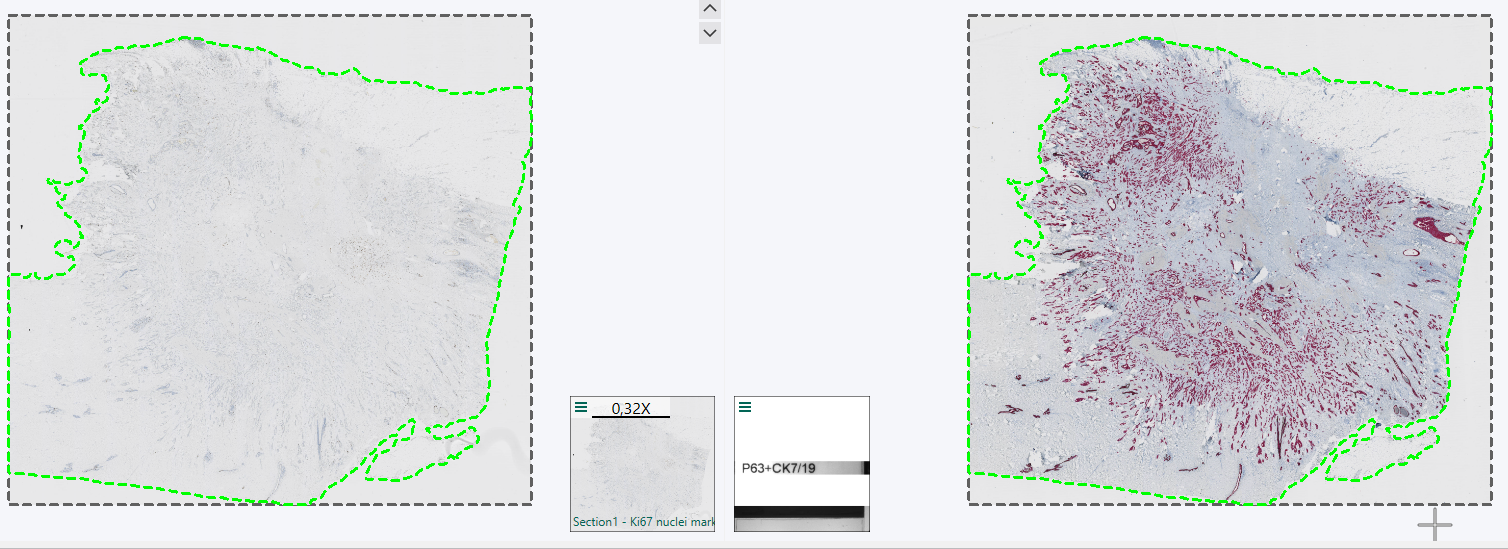
Once the analysis on the aligned Section 2 scan is finalised, and the resulting ROIs, Labels, Heatmaps etc. needs to be exported as layer data, they need to be exported in the Remapped TIFF image Format in order to be aligned with the Section 2 slide. When selecting the Remapped TIFF format after tissue alignment you will be asked which slide the layer data should be aligned to. Here, Section 2 is picked, to remap the ROI layer data from being aligned with Section 1 to Section 2.
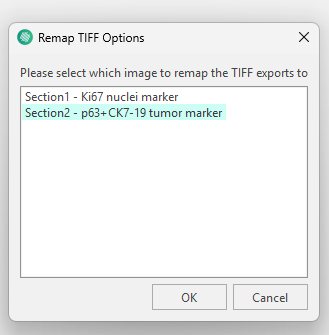
When selecting remapped you will be asked to pick which image to remap on to.
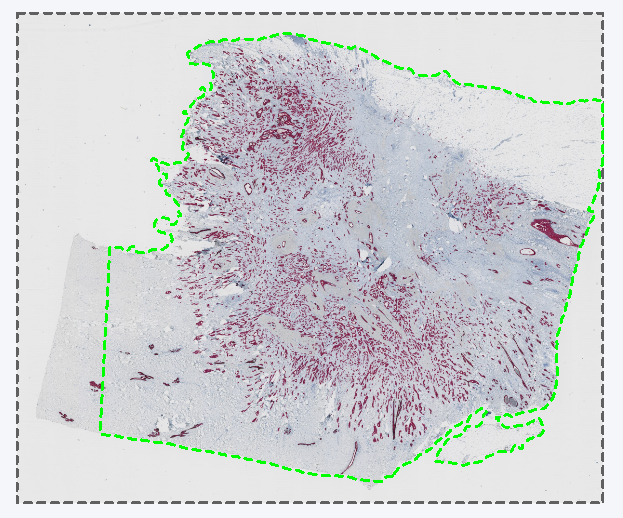
The resulting layer data aligned with the section 2 scan.
Be aware that using the Remapped TIFF format might cause merging issues.
Leica Laser Microdisector (LMD) XML Export
To export Visiopharm ROIs in Leica LMD XML format, ensure you have at least three calibration points. These can be specified by using Visiopharm Text Annotations placed in three well-distributed and easily recognizable image locations. Have the text for the calibration spots being Calibration 1, Calibration 2, ..., as shown in the example image below.
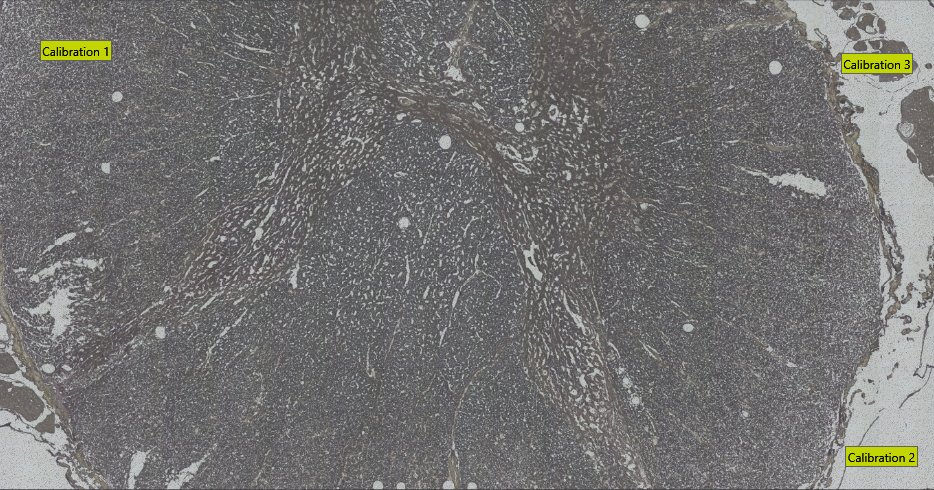
The export will save all non-clear ROI objects on the overlay after performing a morphological closing, followed by a simplification using one-fifth of the closing diameter. The default closing diameter is 50 μm. This can be adjusted in Registry Editor under Computer\HKEY_LOCAL_MACHINE\SOFTWARE\Visiopharm\VPSDK as shown below.
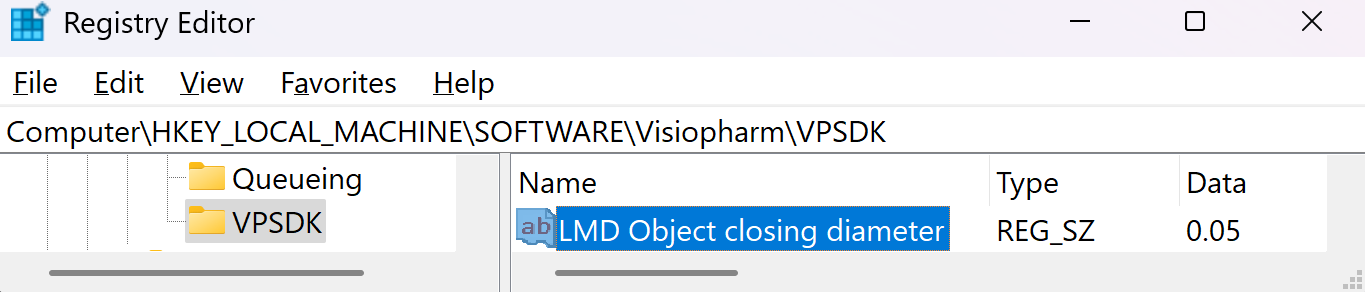
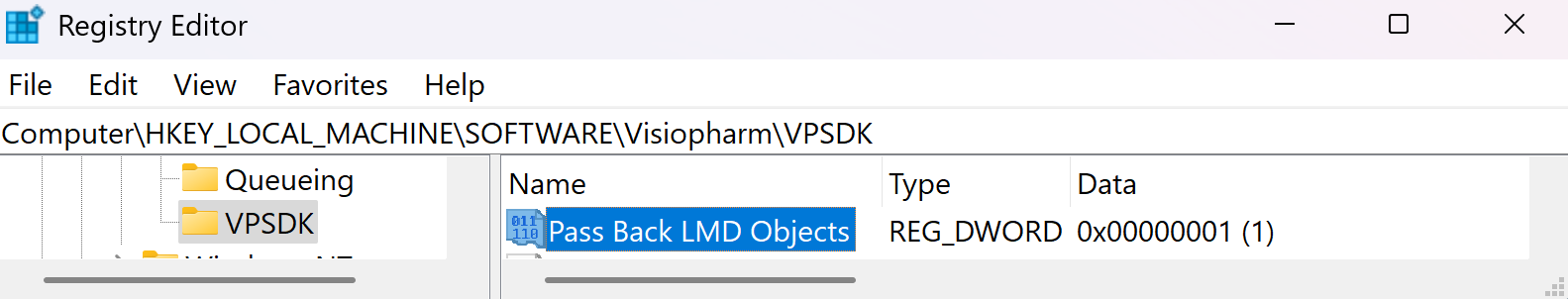
To see which object was written to the LMD xml pass the LMD objects back as annotations to the Visiopharm overlay by enabling the option Pass Back LMD Objects in Registry Editor under Computer\HKEY_LOCAL_MACHINE\SOFTWARE\Visiopharm\VPSDK.
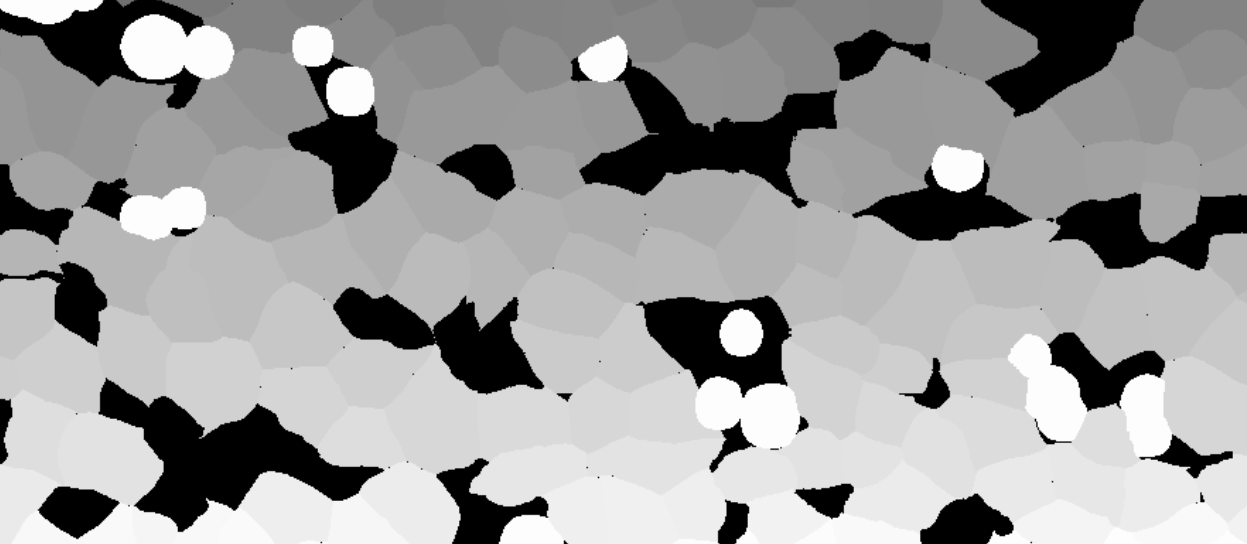
Import of whole slide 32-bit integer value TIFFs
It is possible to import whole slide 32-bit TIFFs with segmentation or annotation data, and convert the objects into Visiopharm Labels, using the Label Crop format. This is neccesary when the TIFF image comes with 32-bit integer values (intensity levels) with uniquely IDed objects. When importing using the Label Crop format, all objects in the imported TIFF file will be converted to overlay Labels of type 1, and objects are split with NN4 (Nearest neighbour) connectivity. 0 intensity will give no Label.
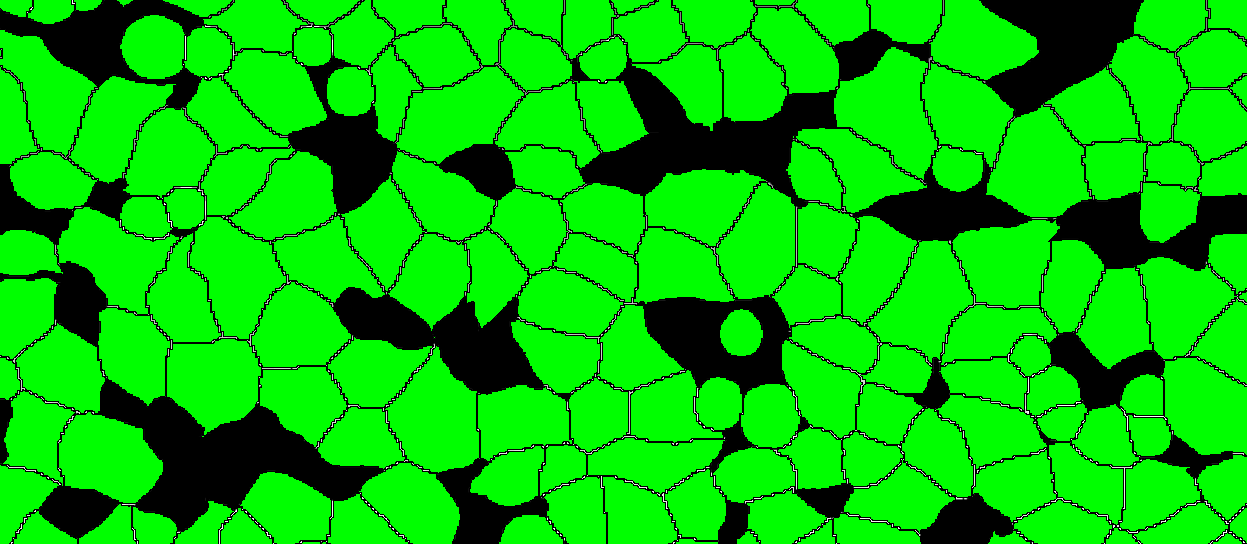
Batch Import Layer Data
-
Click on
 .
. -
Navigate to the folder to import the layer data to.
-
Click on
 and select Layer data to current folder.
and select Layer data to current folder. -
Browse the folder containing the layer data and press Select folder.
-
Select Import folder in the newly opened window.
The layer data files must have a file name matching the file name of the image in the study they should be imported to. This ensures that the files will be imported and attached to the appropriate images.