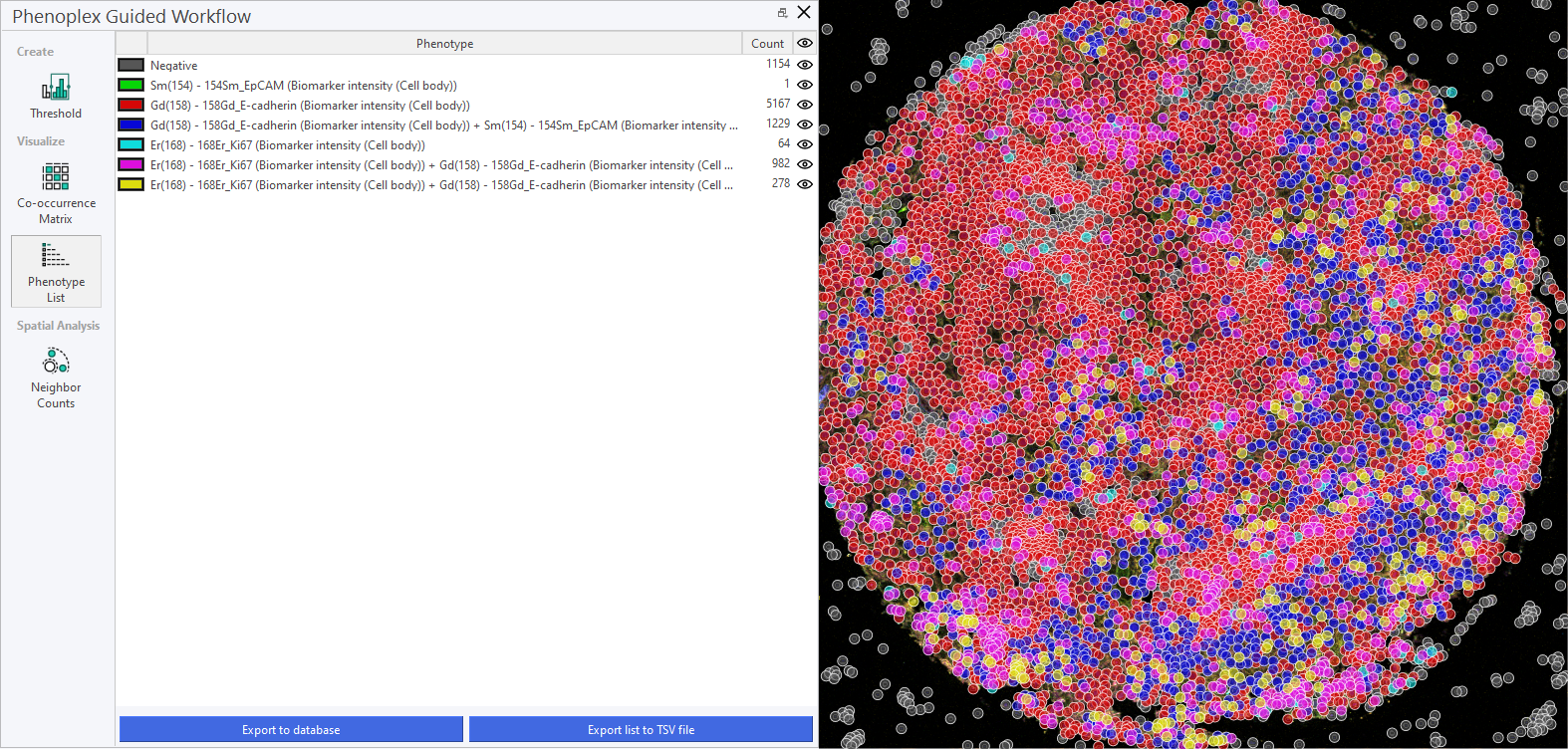
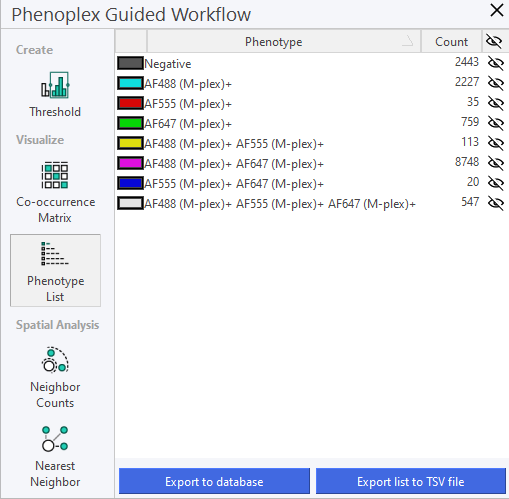
Phenotype List
This section provides a description of the Phenotype List, where identified phenotypes can be managed and reviewed. It serves as a central repository for all phenotypic data. The Phenotype list offers an overview of the selected Variables and their co-occurrences. It also displays the Cell count of the individual phenotypes and their co-occurrences. Visibility can be toggled by clicking on the eye next to the Cell count.
Phenotype Naming and Sorting
This feature introduces a standardized, alphanumeric naming and sorting system for phenotype classification, ensuring that biomarker combinations are displayed in a logical and consistent order. This update simplifies phenotype identification and improves readability, particularly in complex tasks involving multiple biomarkers.
Alphanumeric Naming
Phenotype names are generated by concatenating biomarker names in a strict alphanumeric order, regardless of the functional category or biological significance of each marker. For instance, a phenotype containing CD3, CD8, CD40, and PD-L1 will always be displayed as CD3+ CD8+ CD40+ PD-L1+, rather than reflecting the order in which biomarkers were labeled.
Layered Sorting
Negative Phenotypes: If applicable, these are displayed first in the phenotype list. Single Positives: Phenotypes with single positive markers are listed next, sorted alphanumerically. Double Positives, Triple Positives, etc.: As the complexity of the phenotype increases, phenotypes are sorted in ascending alphanumeric order within each group.
Export phenotypes to Database
Selecting Export to Database adds a new dataset containing all cells and their phenotypes:

Export to TSV file
Clicking on Export list to TSV file will export the phenotype list as a .tsv to any specified folder. This can then be used in the Explore Q/C tool to explore and uncover differences between cohorts.