Phenotype List
This section provides a description of the Phenotype List, where you can manage and review the identified phenotypes. It serves as a central repository for all phenotypic data. The Phenotype list gives you an overview over your selected Variables, and the co-occurrences. It also displays the Cell count of the individual phenotypes and their co-occurrences. You can toggle their visibility by clicking on the eye next to the Cell count.
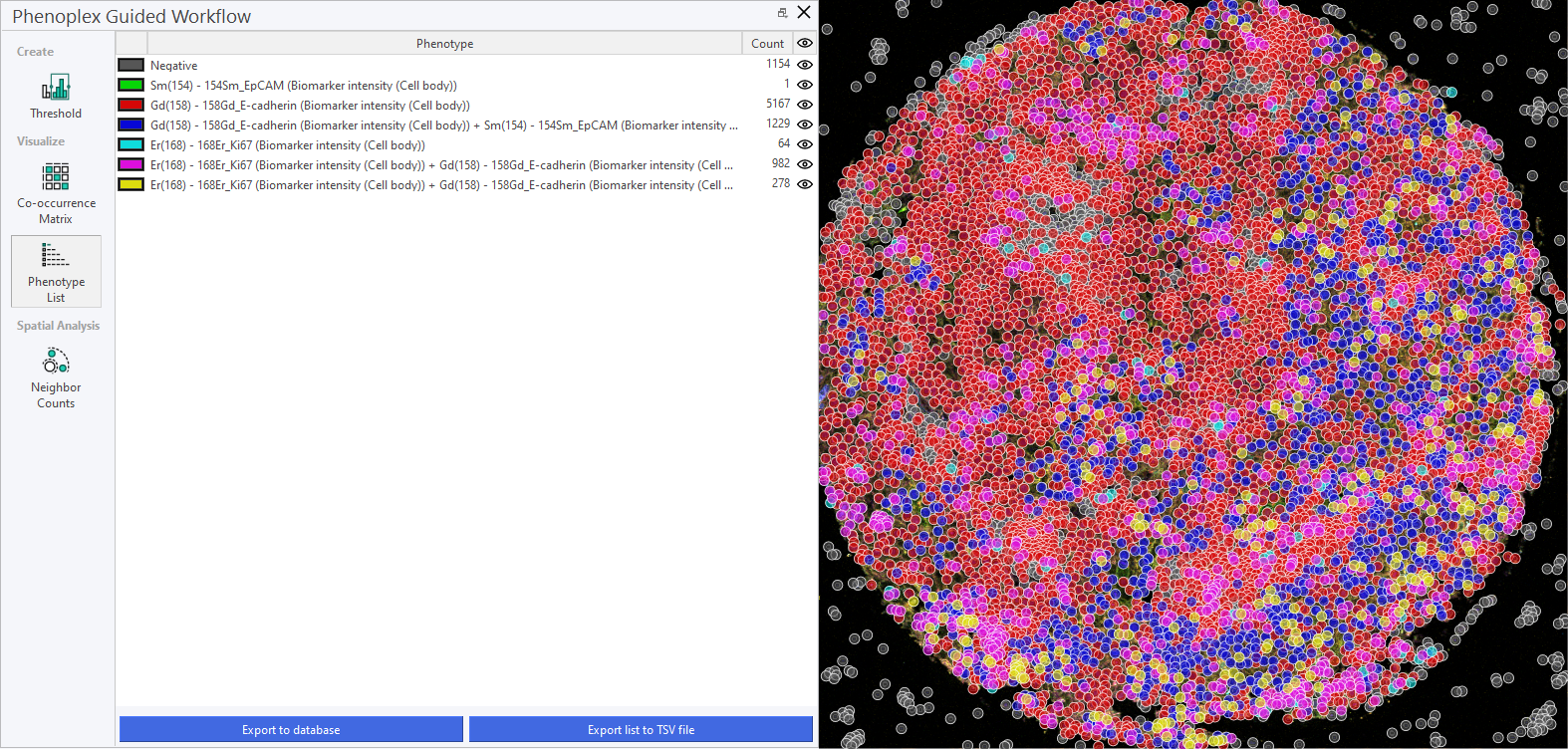
Export phenotypes to Database
When you click on Export to Database a new dataset will be added with all the cells and their phenotypes

Export to TSV file
Clicking on Export list to TSV file will export the phenotype list as a .tsv to any specified folder. This can then be used in the Explore Q/C tool to explore and uncover differences between cohorts.