Semi-automated Phenoplexing
Semi-automated Phenoplexing offers a balance between manual and automated analysis. This approach allows you to apply automated processes where possible, while still retaining manual control for critical decision points. It is ideal for users who need a flexible yet efficient workflow. This guide will take you through the steps for using the semi-automatic phenotyping of cells.
Cell segmentation
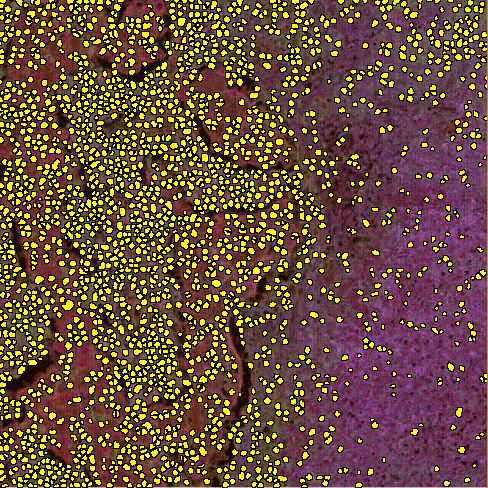
Then the cells should be segmented i.e. the cell objects to include in the automatic phenotyping should be labeled and the labelling should be saved. This can be done manually or by using an APP. In the image below, cells have been segmented and labeled with a single label.
Phenotype APP
Then a clean Phenotype APP should be opened. In the Classification tab setup the contribution from each band in the Channel Contribution Setup dialog that can be opened by pressing the Setup button.
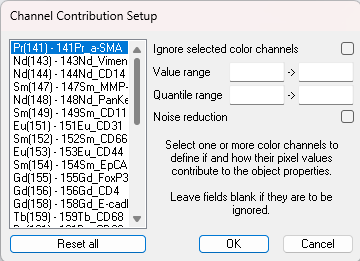
Channel Contribution setup
The Channel Contribution Setup allows for adjustments of the per pixel value input to the automatic calculation of the per object mean value which is the input for the phenotype classifier. If adjustments need to be made it either includes or excludes objects. In this dialog, options regarding which pixels the mean is calculated from are available for each channel/band in the image.
| Option | Description |
|---|---|
| Value range | The value range allows for defining the per pixel value range that will effectively contribute to the calculation of the per object mean value (the input for image classes for output classification algorithm). The low value range input (left) defines a cut-off value by which all pixel values below will be regarded as 0. The upper value range input (right) defines a cut-off value by which all pixel values above will be set to (max value clamping). When no values are entered the data will be treated as raw data (preferable). |
| Quantile range | This quantile range allows disregarding all pixels values outside the range specified by the upper and lower quantile limit. |
| Noise reduction | Enabling the Noise reduction option will run a 3x3 median filter on the pixels. The noise reduction will happen before the value range and quantile range are calculated. |
Adjustments should be made in the following order:
- Change the Quantile range.
- If this is not enough to separate positive and negative cells use the Value range to either reduce the contribution of low signal pixel values and/or reduce the contribution of high value pixels. Avoid the use of Noise reduction when it is not needed (and in case of low positive signal values) as it reduces the applicability of the use of quantiles, which makes it harder to separate noise from signal.
Training the APP
Train the APP by pressing the ![]() Train button. Like other classifiers, training from multiple images is supported. The APP uses all labels, no matter type, currently present in all images for the training.
Train button. Like other classifiers, training from multiple images is supported. The APP uses all labels, no matter type, currently present in all images for the training.
Save the APP
Save the current Phenotype APP as a new APP. This should be done to always ensure having a clean Phenotype APP that has never been trained and thereby changing label names. At this step Post processing steps and output variables could be added.
Run APP on images and save images
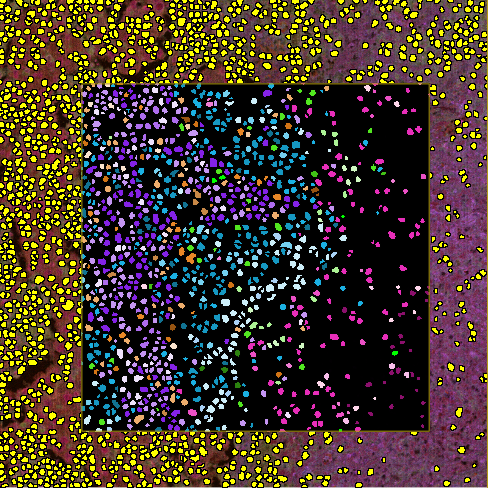
Run the Multiplex Phenotyping APP on slide(s) and save the images. Then, the .csv multiplexing output is saved together with the images in the database. At this point the APP should be ready to use. All label objects from the nuclei segmentation are changed to the respective phenotype label from the phenotype algorithm. Each phenotype is given its own label with a unique color, see Phenotype labeling.